A bioengineer highlights the potential of low-intensity ultrasound for multiple uses, from enhanced drug delivery to the brain to combating cancer


Although lifespan has long been the focus of ageing research, the need to enhance healthspan — the fraction of life spent in good health — is a more pressing societal need. Caloric restriction improves healthspan across eukaryotes but is unrealistic as a societal intervention. Here, we describe the rewiring of a highly conserved nutrient sensing system to prevent senescence onset and declining fitness in budding yeast even when aged on an unrestricted high glucose diet. We show that AMPK activation can prevent the onset of senescence by activating two pathways that remove excess acetyl coenzyme A from the cytoplasm into the mitochondria — the glyoxylate cycle and the carnitine shuttle. However, AMPK represses fatty acid synthesis from acetyl coenzyme A, which is critical for normal cellular function and growth. AMPK activation therefore has positive and negative effects during ageing. Combining AMPK activation with a point mutation in fatty acid synthesis enzyme Acc1 that prevents inhibition by AMPK (the A2A mutant) allows cells to maintain fitness late in life without reducing the mortality associated with advanced age. Our research shows that ageing in yeast is not intrinsically associated with loss of fitness, and that metabolic re-engineering allows high fitness to be preserved to the end of life.
The authors have declared no competing interest.
🚀 Imagine a future where living to 150 is possible—but only if you give up sugar, take ice-cold showers, and inject custom-engineered bacteria. Would you do it? In this deep dive into longevity science, we uncover the shocking truth about genetics, lifestyle, biohacking, and whether living longer actually makes life better. Plus, the real reason happiness might be the ultimate anti-aging hack! 🤯💡
#Longevity #AntiAging #Biohacking #LiveLonger #Science #Health #Wellness #LongevitySecrets #HealthyAging #LifeExtension
University of Queensland researchers are designing nanotechnology they believe could improve how we treat the most aggressive form of breast cancer.
Professor Chengzhong (Michael) Yu and his team are developing novel nanoparticles that could dramatically increase the effectiveness of immunotherapies when treating triple-negative breast cancer (TNBC).
TNBC is aggressive, fast-growing and accounts for 30 per cent of all breast cancer deaths in Australia each year, despite making up only 10 to 15 per cent of new cases.
Professor Yu, from UQ’s Australian Institute for Bioengineering and Nanotechnology (AIBN), said a new solution was needed because TNBC cancer cells lacked the proteins targeted by some of the treatments used against other cancers.
UQ researchers are designing nanotechnology they believe could improve how we treat the most aggressive form of breast cancer.

NASA’s BioNutrients series of experiments is testing ways to use microorganisms to make nutrients that will be needed for human health during future long-duration deep space exploration missions.
Some vital nutrients lack the shelf-life needed to span multi-year human missions, such as a mission to Mars, and may need to be produced in space to support astronaut health. To meet this need, the BioNutrients project uses a biomanufacturing approach similar to making familiar fermented foods, such as yogurt. But these foods will also include specific types and amounts of nutrients that crews will be able to consume in the future.
The first experiment in the series, BioNutrients-1, set out to assess the five-year stability and performance of a hand-held system—called a production pack—that uses an engineered microorganism, yeast, to manufacture fresh vitamins on-demand and in space.

Genetic engineering in non-human primates has long been limited by the need for virus-based gene delivery methods. Recently, researchers in Japan successfully used a nonviral system to introduce a transgene—that is, a gene that has been artificially inserted into an organism—into cynomolgus monkeys, which is a species of primate closely related to humans. The paper is published in the journal Nature Communications.
Small animal models such as mice do not fully replicate the complexity of human diseases, particularly in areas like infectious disease and neuropsychiatric disorders. This limitation has made non-human primates an essential model for biomedical research.
However, genetic modification of these primates has been challenging. For example, conventional virus-based methods require specialized containment facilities and are limited in terms of the size of transgenes that the viruses can carry. Also, these methods do not allow for precise selection of modified embryos before implantation.
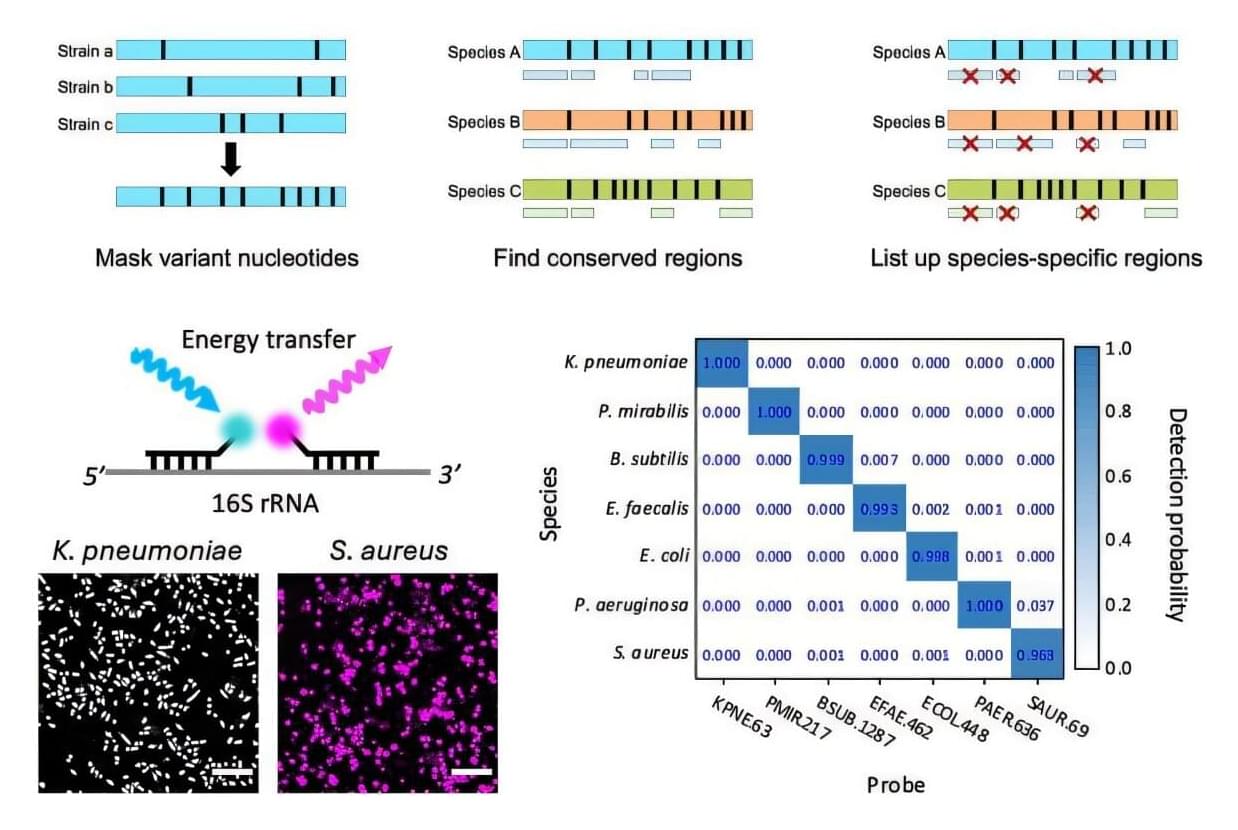
A joint team of professors—Hajun Kim, Taejoon Kwon, and Joo Hun Kang—from the Department of Biomedical Engineering at UNIST has unveiled a novel diagnostic technique that utilizes artificially designed polymers known as peptide nucleic acid (PNA) as probes. The research is published in the journal Biosensors and Bioelectronics.
The fluorescence in situ hybridization (FISH) technique works by detecting fluorescent signals generated when probe molecules bind to specific genetic sequences in bacteria. This innovative FISH method employs two PNA molecules simultaneously. By analyzing the genomic sequences of 20,000 bacterial species, the research team designed PNA sequences that specifically target the ribosomal RNA of particular species.
The method is significantly faster and more accurate than traditional bacterial culture and polymerase chain reaction (PCR) analysis, and it holds promise for reducing mortality rates in critical conditions such as sepsis, where timely administration of antibiotics is crucial.
Professor Kwang-Hyun Cho’s research team has recently been highlighted for their work on developing an original technology for cancer reversal treatment that does not kill cancer cells but only changes their characteristics to reverse them to a state similar to normal cells. This time, they have succeeded in revealing for the first time that a molecular switch that can induce cancer reversal at the moment when normal cells change into cancer cells is hidden in the genetic network.
KAIST (President Kwang-Hyung Lee) announced on the 5th of February that Professor Kwang-Hyun Cho’s research team of the Department of Bio and Brain Engineering has succeeded in developing a fundamental technology to capture the critical transition phenomenon at the moment when normal cells change into cancer cells and analyze it to discover a molecular switch that can revert cancer cells back into normal cells.
A critical transition is a phenomenon in which a sudden change in state occurs at a specific point in time, like water changing into steam at 100℃. This critical transition phenomenon also occurs in the process in which normal cells change into cancer cells at a specific point in time due to the accumulation of genetic and epigenetic changes.

What makes us unique? Different from most, yet similar to a few? What shapes our physical, behavioral, and even mental makeup? The answer lies in our genes.
Passed from parents to their offspring, genes contain the information that specifies physical and biological traits.
But that’s not all. Genes are also responsible for diseases. Faulty genes can cause all kinds of issues that can manifest as birth defects, chronic diseases, or developmental problems.