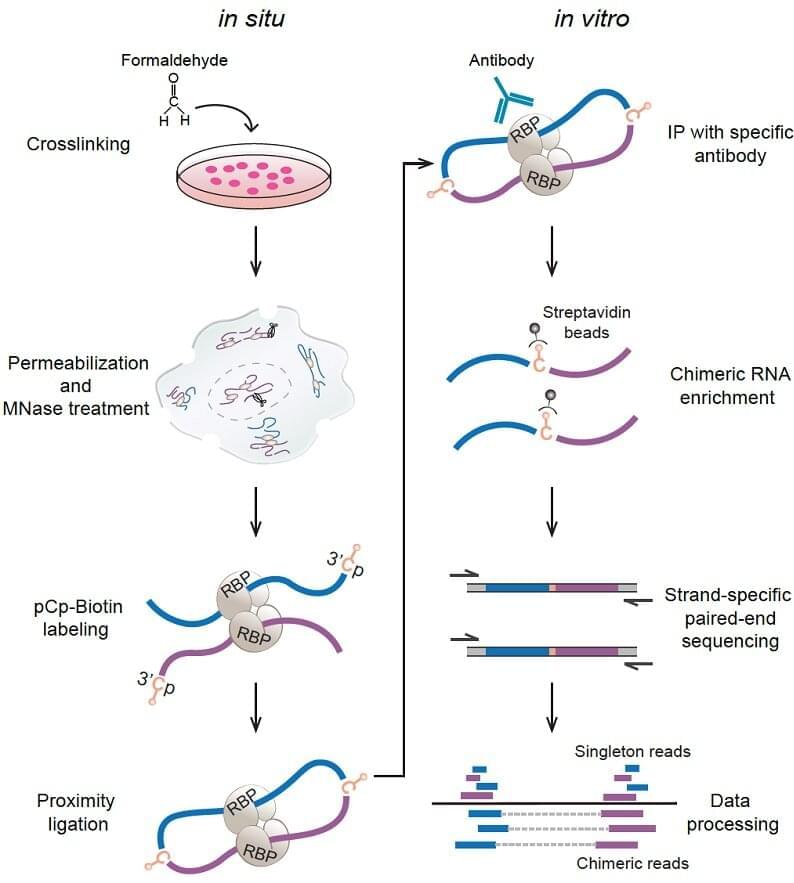
A research team led by Prof. Xue Yuanchao from the Institute of Biophysics of the Chinese Academy of Sciences has developed a new method for global profiling of in-situ RNA–RNA contacts associated with a specific RNA-binding protein (RBP) and revealed positional mechanisms by which PTBP1-associated RNA loops regulate cassette exon splicing.
This study was published online in Molecular Cell on March 22.
In eukaryotes, the same pre-mRNA can produce multiple protein isoforms to execute similar or different biological functions through alternative splicing. Several longstanding models proposed that RBPs may regulate alternative splicing by modulating long-range RNA–RNA interactions (RRI). However, direct experimental evidence was lacking.