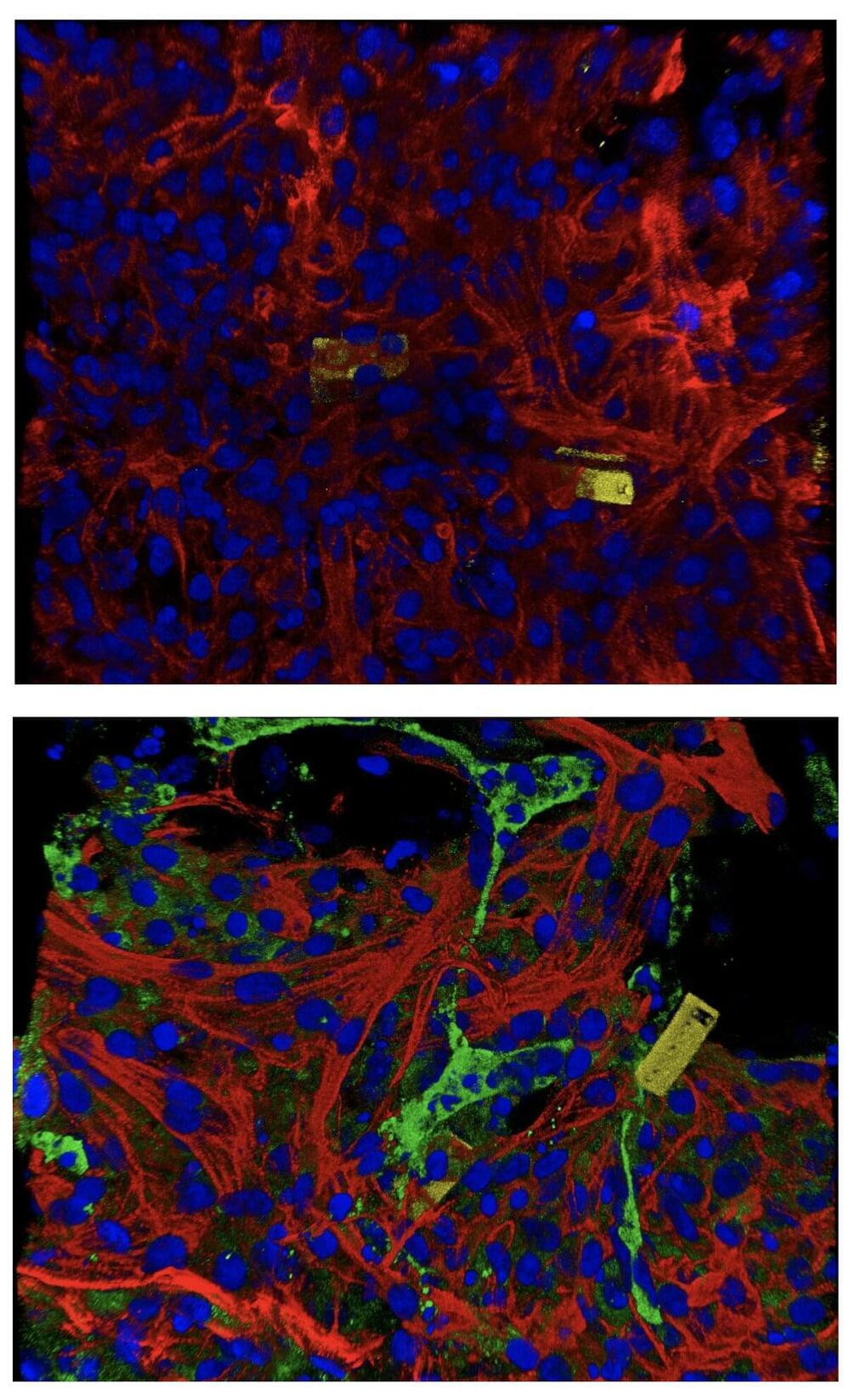
Making living cells blink fluorescently like party lights may sound frivolous. But the demonstration that it’s possible could be a step toward someday programming our body’s immune cells to attack cancers more effectively and safely.
That’s the promise of the field called synthetic biology. While molecular biologists strip cells down to their component genes and molecules to see how they work, synthetic biologists tinker with cells to get them to perform new feats — discovering new secrets about how life works in the process. In this episode, Steven Strogatz talks with Michael Elowitz, a professor of biology and bioengineering at the California Institute of Technology and a Howard Hughes Medical Institute Investigator.