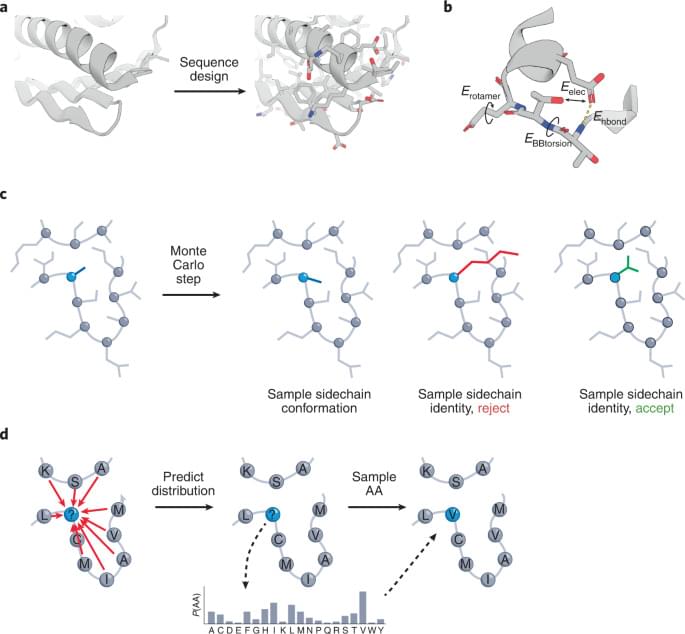
The design of protein sequences that can precisely fold into pre-specified 3D structures is a challenging task. A recently proposed deep-learning algorithm improves such designs when compared with traditional, physics-based protein design approaches.
ABACUS-R is trained on the task of predicting the AA at a given residue, using information about that residue’s backbone structure, and the backbone and AA of neighboring residues in space. To do this, ABACUS-R uses the Transformer neural network architecture6, which offers flexibility in representing and integrating information between different residues. Although these aspects are similar to a previous network2, ABACUS-R adds auxiliary training tasks, such as predicting secondary structures, solvent exposure and sidechain torsion angles. These outputs aren’t needed during design but help with training and increase sequence recovery by about 6%. To design a protein sequence, ABACUS-R uses an iterative ‘denoising’ process (Fig.